This is a Mastodon thread. The original thread is available here:
🚨 Publication alert! 🚨
At EHPS 2021, Szilvia Zörgő presented “Bridging the qualitative and quantitative with Epistemic Network Analysis: a worked example”. 🌉
Based on this, we were invited to write a tutorial for the Advanced Methods section of an EHPS #OpenAccess journal, Health Psychology and Behavioral Medicine.
That tutorial is now out!
Get it at https://doi.org/jrfb - main points in this 🧵!
#Psychology #OpenScience #BehaviorChange #QualitativeMethods #RStats @rstats@gup.pe
🧵 1/21
Qualitative, quantitative, and ENA
Qualitative methods are typically contrasted with quantitative methods – so much so that at EHPS 2012, the Synergy Expert Meeting addressed overcoming the ‘irreconcilable epistemological differences’ between both approaches.
Analyses have described this contrast using terms such as facts vs opinions, objective versus subjective, and masculine versus feminine.
🧵 2/21

If qualitative and quantitative methods are combined within one study (i.e., a mixed-methods study), they often have complementary functions.
Epistemic Network Analysis is a method that instead strives to unify qualitative and quantitative approaches, ultimately producing data visualizations that provide a novel lens to aid understanding the patterns in qualitative datasets.
🧵 3/21
ROCK to ENA
In this tutorial, we explain how to create an Epistemic Network for a qualitative dataset.
Because we want to work conform the #OpenScience principles, we use the Reproducible Open Coding Kit (the ROCK 🤘).
The ROCK is an open standard for coded qualitative data, and we also explain a bit about that.
🧵 4/21

The example study
As illustration, we use a qualitative study into perceptions of illness etiology, comparing patients who chose complementary or alternative medicine with patients who chose biomedical treatments. This example uses five codes:
1️⃣ ‘psych’: emotion, stress, trauma, nerves
2️⃣ ‘vital’: energy/qi/prana, flow, block
3️⃣ ‘eco’: environmental toxins, chemicals
4️⃣ ‘nutri’: quality or type of food, additives
5️⃣ ‘genetic’: inherited illness or susceptibility, genes
🧵 5/21

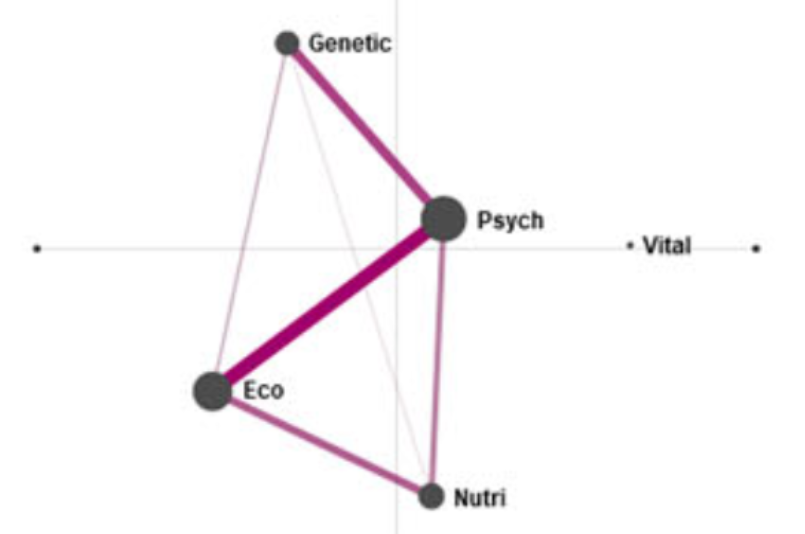
This figure shows Epistemic Networks from this example. The left (purple) network is for patients who chose biomedicine, the right (green) network is for patients who chose complementary or alternative medicine, and the center network shows the difference between the other two plots.
The edges represent how often codes co-occurred: for example, in the biomedicine group, the “psych” and “genetic” codes co-occurred most frequently. This allows easy visual comparison of such patterns.
🧵 6/21
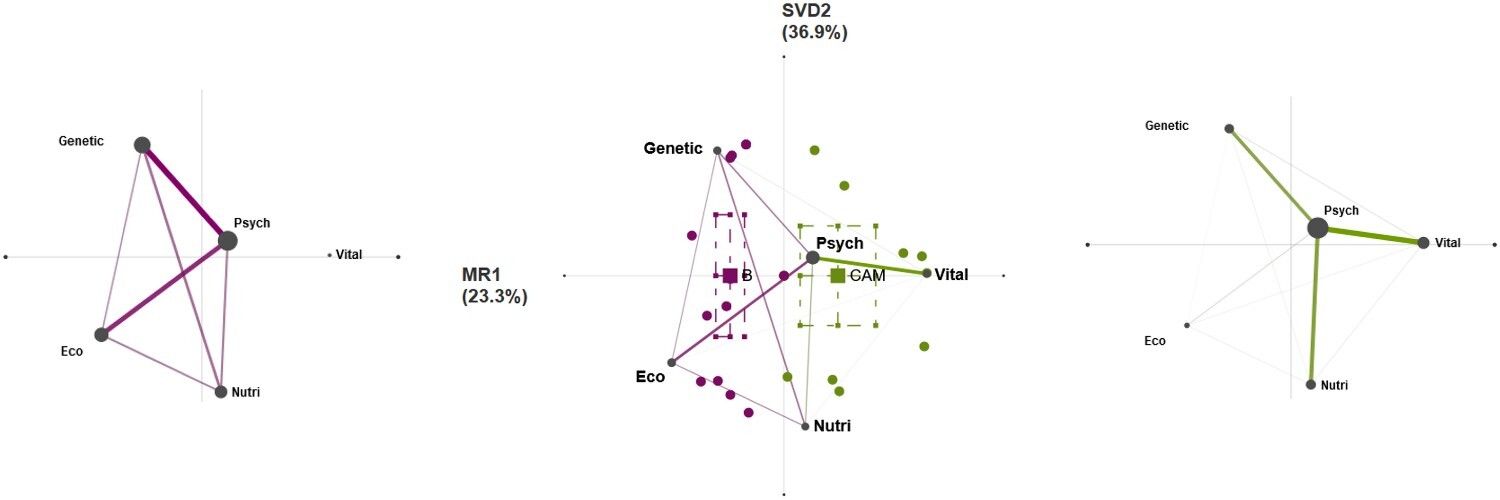
But that is not all! With ENA, you can create networks for any subsample of participants! 🤩
For example, these are six networks also split on diagnosis (columns). This shows that there is considerably heterogeneity within each diagnosis group, placing the differences between patients choosing the biomedical route and those choosing alternative of complementary medicine in perspective. 👓
🧵 7/21
The CLIFF
So, next we explain how you can obtain those visualizations!
Before we begin: in the tutorial, we follow the Clean Lean Initial Formatted File (CLIFF).
That’s a prestructured set of directories and files meant to help you start a ROCK project. It’s available in a Git repository at https://rock.science/cliff.
Don’t worry – you don’t need Git to use the CLIFF – you can also download as a .ZIP file!
🧵 8/21

The ROCK standard
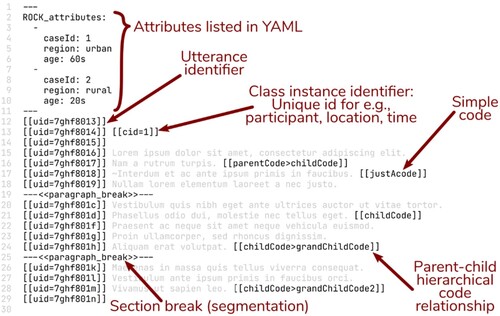
The ROCK standard is designed as a human- and machine-readable standard for coded qualitative data. Some terms:
👉
source: a file containing qualitative data👉
utterance: smallest codable unit of data, typically (but not necessarily) a sentence👉
identifier: a character string added to the data to represent codes and other things👉
code: a concept/construct of interest. Codes are represented by their code identifier in double brackets:[[code]],[[parent>child]]
🧵 9/21

👉
section break: a type of identifier used to segment data, by inserting a line containing e.g.---<<section_break_id>>---👉
class: a group, e.g. cases (participants) or interview locations👉
class instance: a single case or location:[[cid=1]],[[locationId=library]]👉
attribute: characteristics of class instances (e.g. age, diagnosis, etc), attached to utterances through class instance identifiers👉
utterance identifier: a unique identifier for an utterance
🧵 10/21
The {rock} R 📦
To work with these ROCK files, we will use the {rock} R package. You can get it from CRAN using:
install.packages('rock');
You can use the ROCK standard without using the {rock} R package, but as yet, the standard hasn’t been implemented in any other software yet that we know of. In addition, the {rock} R package produces a .CSV file that we can import into the rENA tool to create the Epistemic Networks.
🧵 11/21
Step-by-step tutorial
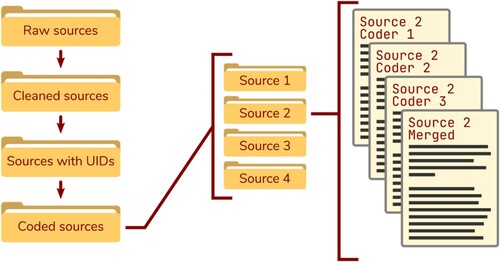
We place our raw data sources as plain text files in a directory named 010—raw-sources.
Usually raw sources are a bit messy, and the ROCK requires every utterance to be on a separate line. For this, we use the rock::clean_sources() function.
As its input argument, we specify the directory with the raw sourcs, and as its output argument, the directory where we want to write the clean sources 020—cleaned-sources.
🧵 12/21

Next, we add utterance identifiers before every line. These uniquely identify every utterance, which makes two things possible.
1st, efficient reference to utterances, e.g. to add codes or describe results.
2nd, when multiple coders code the same source independently, utterance identifiers allow determining which codes were attached to each utterance (and merging them).
To prepend utterance identifiers, use rock::prepend_ids_to_sources(), again specifying the input and output paths.
🧵 13/21
Attributes are specified for each class instance. The most common type of class is participants (or ‘cases’, following the REFI terms), so each instance or case is a person. In the YAML chunk, for each class instance identifier, attributes can be specified - this enables querying data and codes using those attributes.
Attributes are indicated using YAML fragments. This is a bit hard to explain in a toot, but feel free to peruse the article 😬
🧵 14/21

Coding
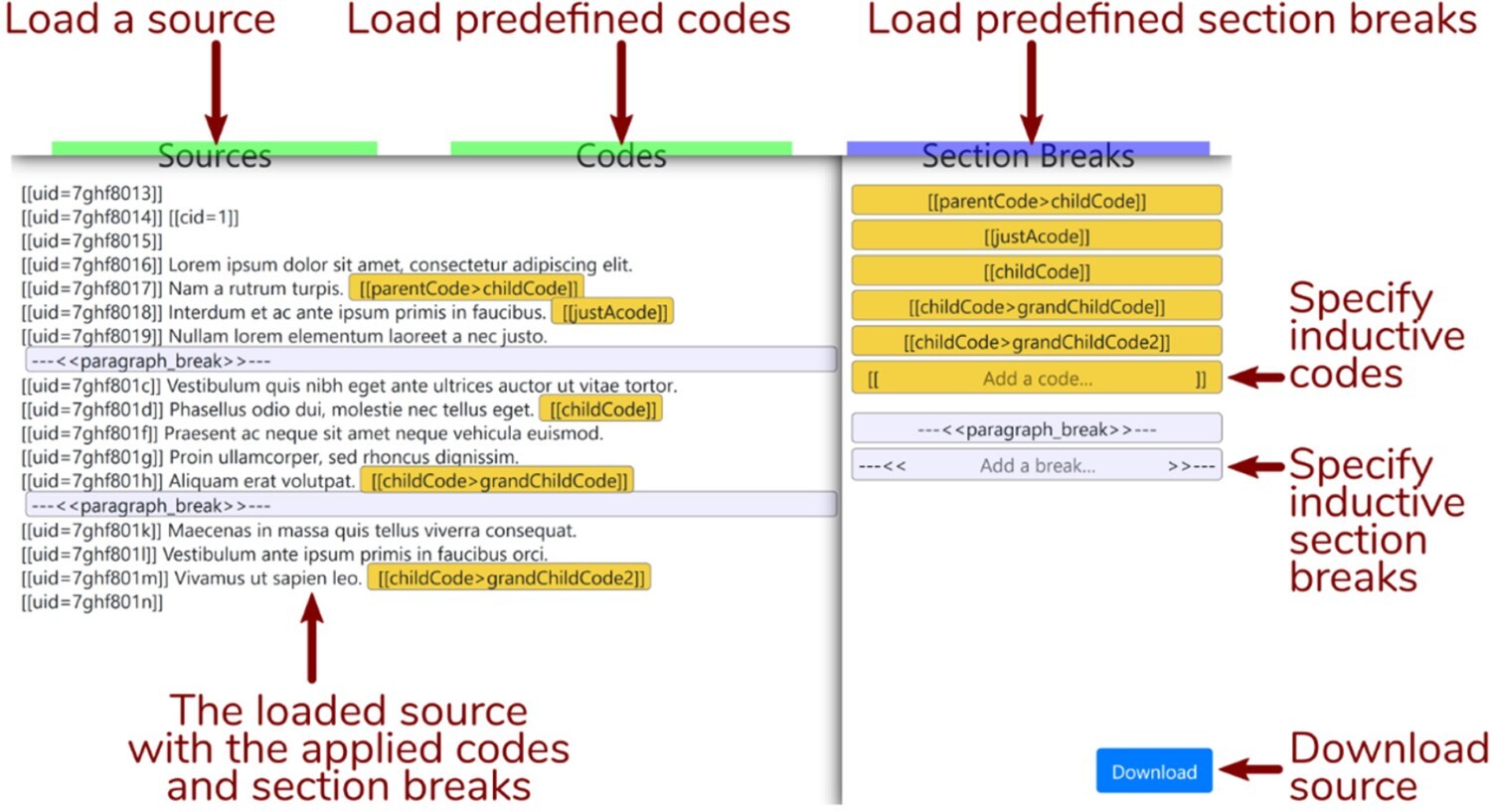
Coding qualitative data in the ROCK standard just consists of adding code identifiers to the plain text sources.
This can be done with any text editor, but there is also a rudimentary (but GDPR-compliant) javascript/html/css web app available at https://i.rock.science.
It’s very simple: just load a source, create some codes (or load those from a file, too) and you can drag and drop the codes to code utterances.
🧵 15/21
Merging codes from independent coders
If you used independent coders, the utterance identifiers make it possible to merge the codes from all coders into one primary source.
The {rock} R 📦 has a function for this: rock::merge_sources(). You can specify which ones are the primary sources by regular expression or by path.
This writes new sources to disk that contain the codes from all separate sources (using the utterance identifiers).
🧵 16/21
Parsing the coded sources
If you got to this point (in the tutorial in the paper, not this tootorial 😬), you have coded and merged sources, so we’re ready to parse them.
In the {rock} R 📦, there’s rock::parse_sources() function for this. It produces an object with a lot of info, but the only thing we need for ENA is the mergedSourceDf data frame, which we can export with the rock::export_mergedSourceDf_to_csv() function.
That .csv file can then be imported into the ENA web app to produce Epistemic Networks.
🧵 17/21
Epistemic Network Analyses
To create an Epistemic Network, you can use the web app available at https://app.epistemicnetwork.org.
You have to specify which column designates ‘units’, which designates ‘conversations’, and which contain the codes you want to see in the network. In ENA, co-occurrences are computed per ‘unit’, and only for codes from the same ‘conversation’. So, ‘conversations’ are usually interviews (e.g., sources), and units are often participants (or groups of participants).
🧵 18/21
You also have to specify “stanza window”, which also determines when codes are considered to co-occur. For more details on the meaning of ‘units’, ‘conversations’, and ‘stanzas’, as well as how the networks are created from the coded utterances, please see the tutorial at https://doi.org/jrfb 🙂
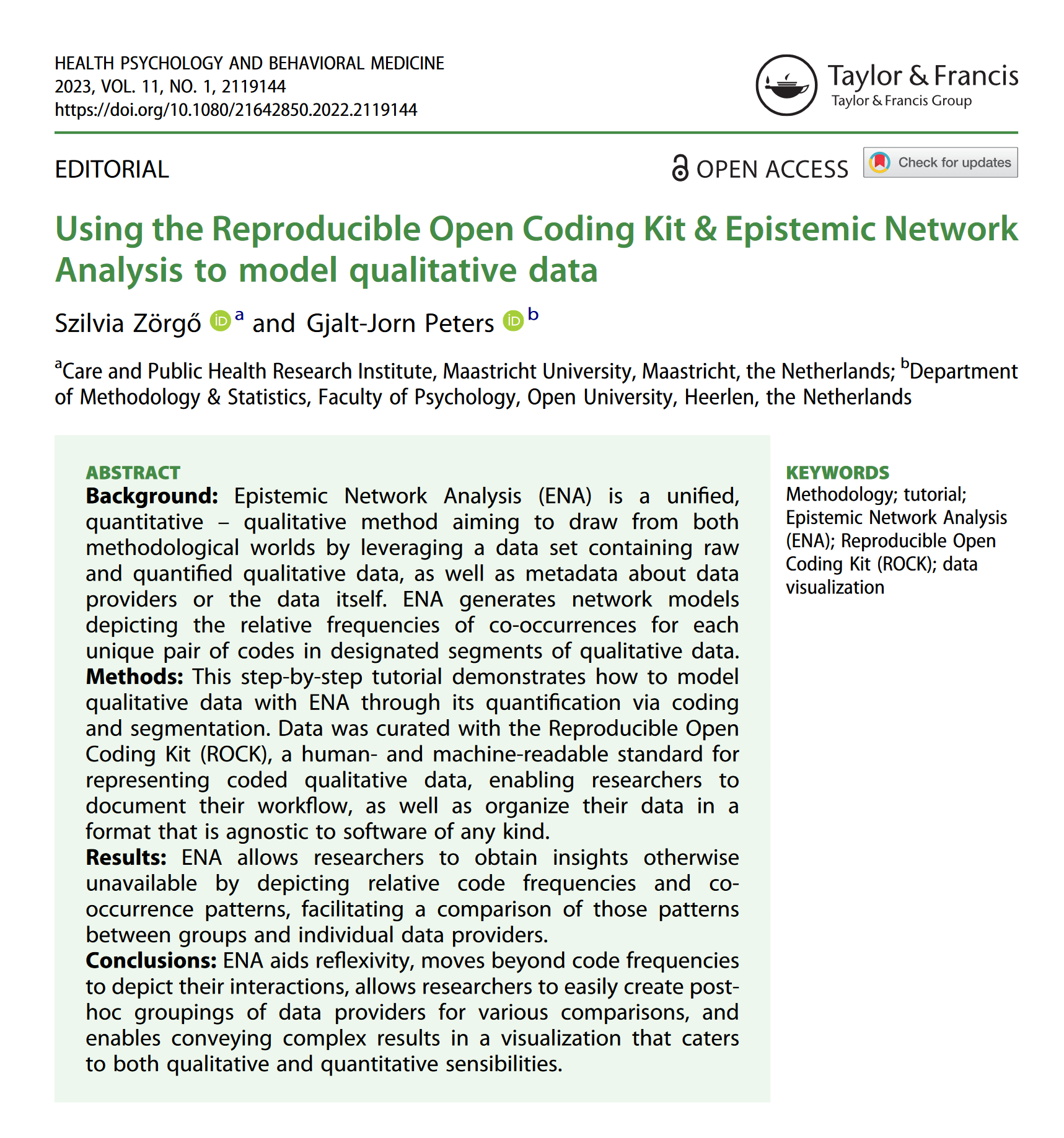
You should now be rewarded with a pretty visualization as shown here 🤩
Well done! 💪
🧵 19/21

Conclusion
In the tutorial, we also discuss a number of strengths and limitations of Epistemic Network Analysis.
ENA offers a unique perspective on your data, helping you see patterns you may otherwise miss. By inspecting the networks in parallel with the coded data, you can use the richness of qualitative data to its fullest, while also benefiting from the visualizations possible by quantification.
🧵 20/21
In the process we introduced the Reproducible Open Coding Kit, a human- and machine-readable standard for coded qualitative data. We also showed some functions from the {rock} R 📦 to help you work with .rock files.
We hope this tutorial can help you to answer new, interesting research questions - and in an Open Science way!
Oh, and the tutorial itself was at https://doi.org/jrfb 🙂
🧵 21/21
